Statistics of the predicted circRNAs
This database provide the first largest profiling of tissue and stress-specific circRNAs in tea or any single plant species. From 5 different tissues of TV-1 genotype, a total of 3,052 full length circRNA isoforms were predicted and from downloaded RNA-seq data of tea from NCBI, a total of 59,575 full-length circRNAs were identified.
Among 59,575 circRNAs, 25,651 were detected form stress-related data and 39,750 circRNAs from tissue-related data with only 5,826 circRNAs common between these two sets of data. The length of these circRNAs in the database ranges from 26 to 19,157 nucleotides with upto maximum of 7 circexons and 4201 BSJ read counts.
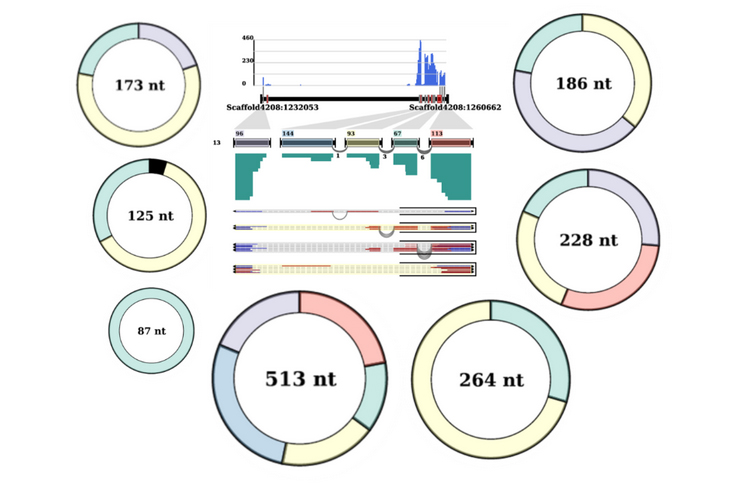
Please go through our paper for details on how we predict full-length cicrRNAs

