Which data we used?
We have generated RNA-seq data using 5 different tissues of TV-1 genotype of Indian tea (Camellia assamica) for the de-novo prediction of circRNAs and differential expression analysis to identify tissue specific circRNAs.
Moreover we found that 81 paired-end RNA-seq data for different stress-related conditions and 146 belonging to 17 different tissues of tea (Camellia sinensis) is available at NCBI. We have downloaded these paired-end RNA-seq data and used for the in-silico prediction of circRNAs.
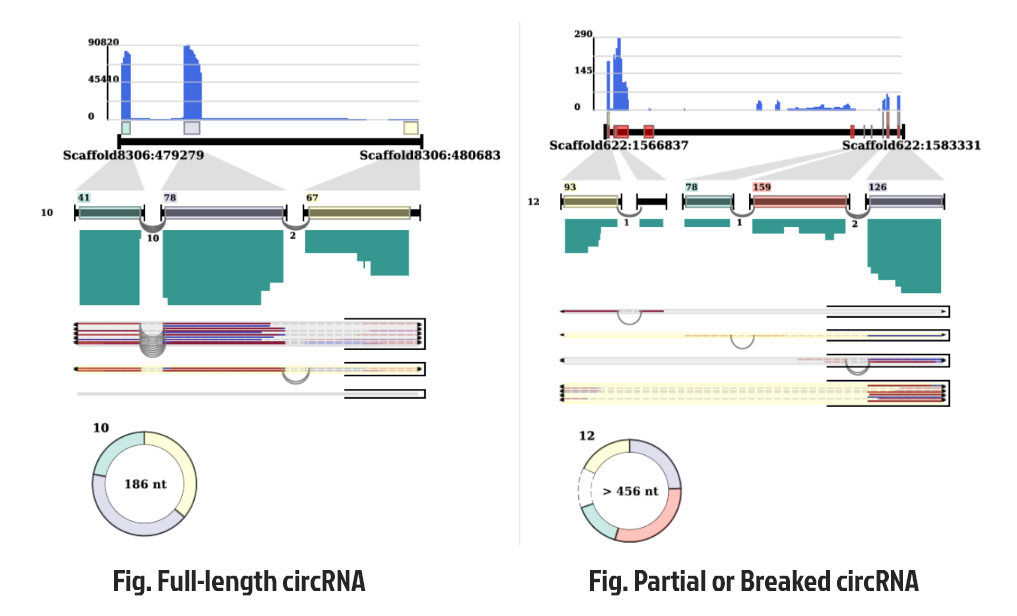
Please go through our paper for details on how we predict full-length cicrRNAs

